Code
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as pltkakamana
January 7, 2023
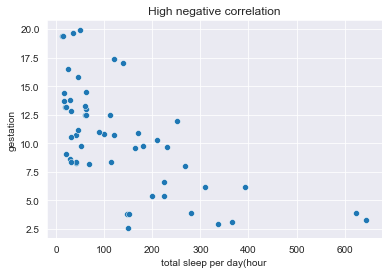
In this article we will explore basically a linear relationship between two variables, its possible quantification (magnitude & direction). We will also touch high level of confounding & caveats of correlation. This article use exploration of study for mammals sleeping habits & world happiness
This is my learning experience of data science through DataCamp
| species | body_wt | brain_wt | non_dreaming | dreaming | total_sleep | life_span | gestation | predation | exposure | danger | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Africanelephant | 6654.000 | 5712.0 | NaN | NaN | 3.3 | 38.6 | 645.0 | 3 | 5 | 3 |
| 1 | Africangiantpouchedrat | 1.000 | 6.6 | 6.3 | 2.0 | 8.3 | 4.5 | 42.0 | 3 | 1 | 3 |
| 2 | ArcticFox | 3.385 | 44.5 | NaN | NaN | 12.5 | 14.0 | 60.0 | 1 | 1 | 1 |
| 3 | Arcticgroundsquirrel | 0.920 | 5.7 | NaN | NaN | 16.5 | NaN | 25.0 | 5 | 2 | 3 |
| 4 | Asianelephant | 2547.000 | 4603.0 | 2.1 | 1.8 | 3.9 | 69.0 | 624.0 | 3 | 5 | 4 |
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 62 entries, 0 to 61
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 species 62 non-null object
1 body_wt 62 non-null float64
2 brain_wt 62 non-null float64
3 non_dreaming 48 non-null float64
4 dreaming 50 non-null float64
5 total_sleep 58 non-null float64
6 life_span 58 non-null float64
7 gestation 58 non-null float64
8 predation 62 non-null int64
9 exposure 62 non-null int64
10 danger 62 non-null int64
dtypes: float64(7), int64(3), object(1)
memory usage: 5.5+ KBThe sleep time of 39 species of mammals distributed over 13 orders is analyzed in regards to their distribution over the 13 orders. There are 62 observations across 11 variables.
species : Mammal species
body_wt : Mammal’s total body weight (kg)
brain_wt : Mammal’s brain weight (kg)
non_dreaming : Sleep hours without dreaming
dreaming : Sleep hours spent dreaming
total_sleep : Total number of hours of sleep
life_span : Life span (in years)
gestation : Days during gestation / pregnancy
The likelihood that a mammal will be preyed upon. 1 = least likely to be preyed on. 5 = most likely to be preyed upon.
exposure : How exposed a mammal is during sleep. 1 = least exposed (e.g., sleeps in a well-protected den). 5 = most exposed.
A measure of how much danger the mammal faces. This index is based upon Predation and Exposure. 1 = least danger from other animals. 5 = most danger from other animals.
<AxesSubplot:title={'center':'Mammals Body Weight Distribution'}, xlabel='body_wt', ylabel='Count'>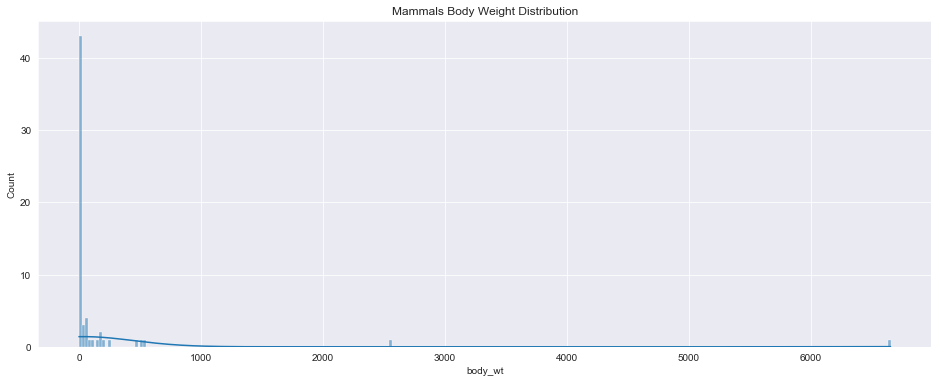
<AxesSubplot:title={'center':'Top 10 Mammals Body Wight'}, xlabel='body_wt', ylabel='species'>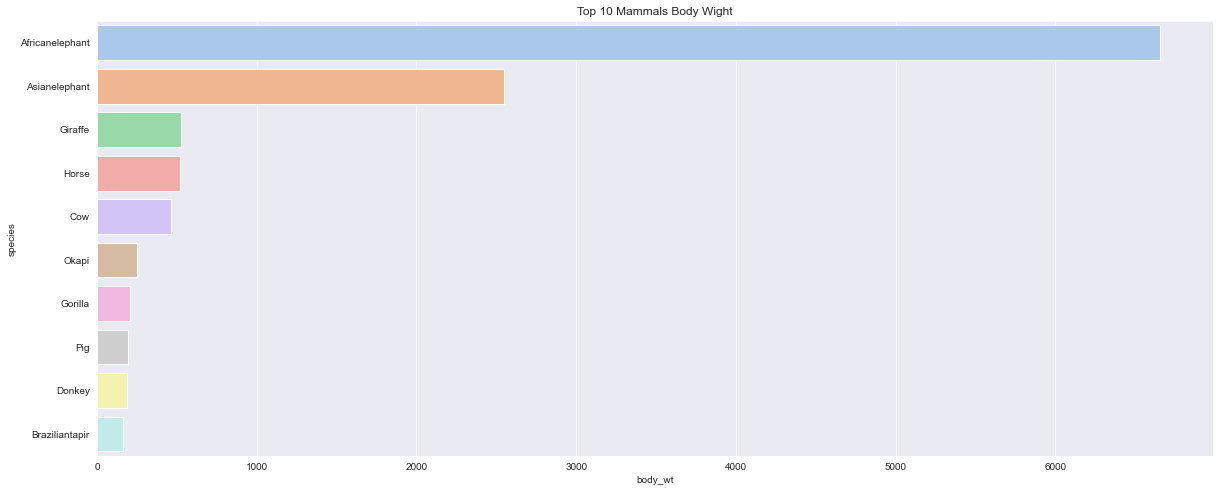
<AxesSubplot:title={'center':'Mammals Brain Weight Distribution'}, xlabel='brain_wt', ylabel='Count'>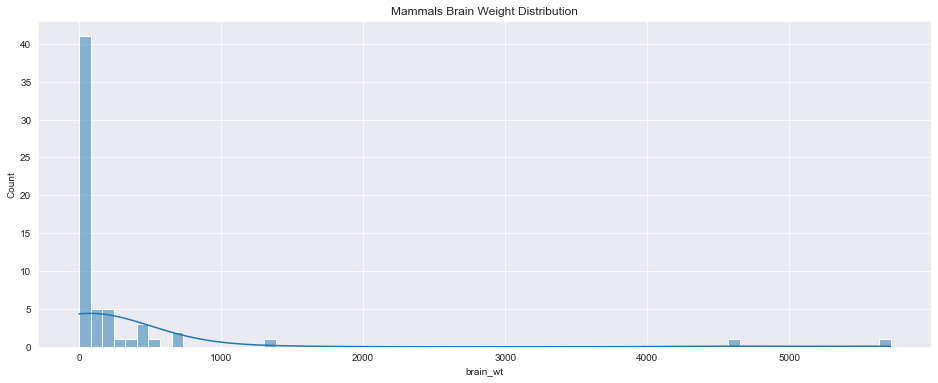
<AxesSubplot:title={'center':'Top 10 Mammals Brain Wight'}, xlabel='brain_wt', ylabel='species'>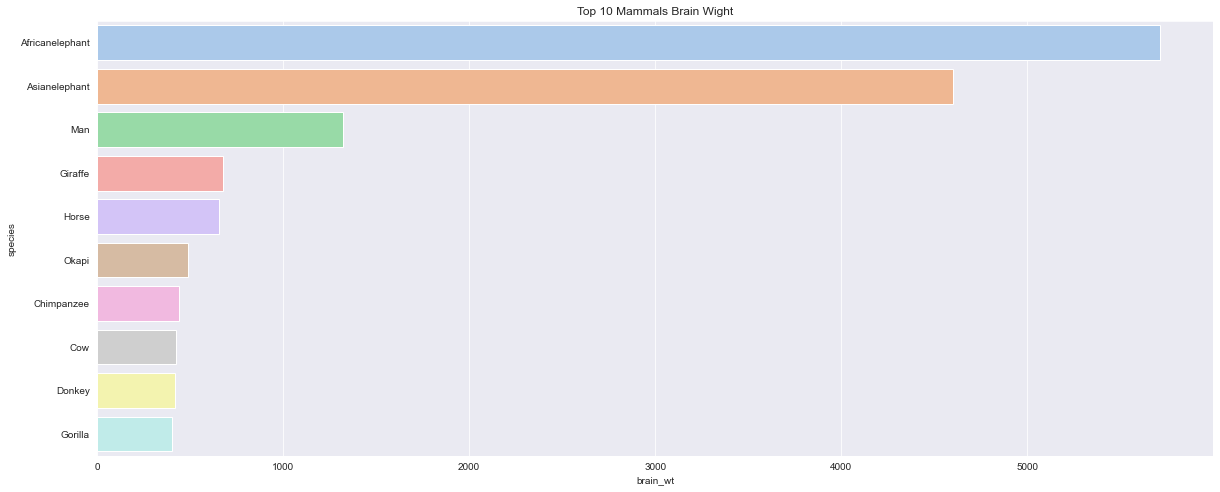
<AxesSubplot:title={'center':'Mammals Life Span Distribution'}, xlabel='life_span', ylabel='Count'>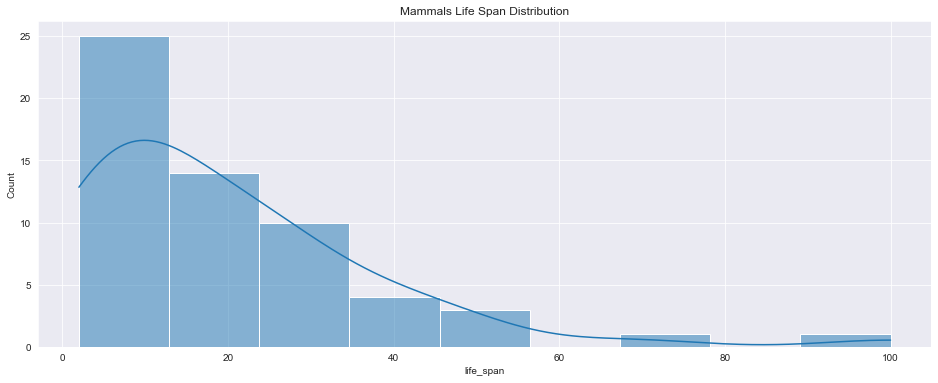
<AxesSubplot:title={'center':'Top 10 Mammals Life Span'}, xlabel='life_span', ylabel='species'>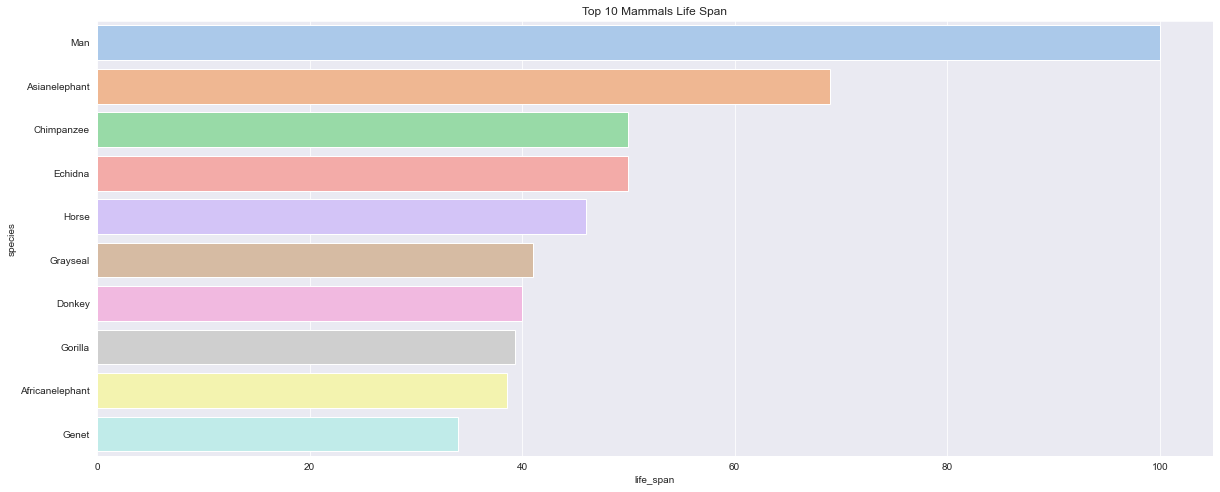
figure, axes = plt.subplots(2, 2, sharex=True, figsize=(18,10))
figure.suptitle('Predation Total Sleep Visualization')
sns.countplot(x='predation',data=df,palette='pastel', ax=axes[0][0])
sns.boxplot(x="predation", y="total_sleep", data=df, palette='pastel', ax=axes[0][1])
sns.violinplot(x="predation", y="total_sleep", data=df, palette='pastel', ax=axes[1][0])
sns.stripplot(x="predation", y="total_sleep", data=df,jitter=True, palette='pastel', ax=axes[1][1])C:\Users\dghr201\AppData\Local\Temp\ipykernel_42120\3907231976.py:6: FutureWarning: Passing `palette` without assigning `hue` is deprecated.
sns.stripplot(x="predation", y="total_sleep", data=df,jitter=True, palette='pastel', ax=axes[1][1])<AxesSubplot:xlabel='predation', ylabel='total_sleep'>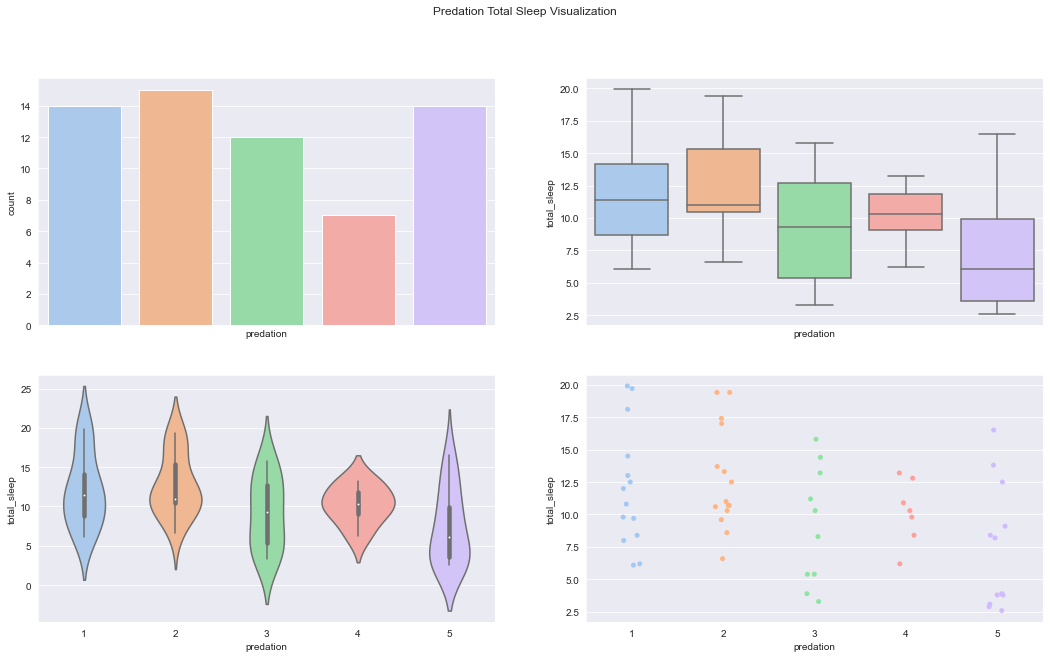
figure, axes = plt.subplots(2, 2, sharex=True, figsize=(18,10))
figure.suptitle('Exposure Total Sleep Visualization')
sns.countplot(x='exposure',data=df,palette='pastel', ax=axes[0][0])
sns.boxplot(x="exposure", y="total_sleep", data=df, palette='pastel', ax=axes[0][1])
sns.violinplot(x="exposure", y="total_sleep", data=df, palette='pastel', ax=axes[1][0])
sns.stripplot(x="exposure", y="total_sleep", data=df,jitter=True, palette='pastel', ax=axes[1][1])C:\Users\dghr201\AppData\Local\Temp\ipykernel_42120\3542283944.py:6: FutureWarning: Passing `palette` without assigning `hue` is deprecated.
sns.stripplot(x="exposure", y="total_sleep", data=df,jitter=True, palette='pastel', ax=axes[1][1])<AxesSubplot:xlabel='exposure', ylabel='total_sleep'>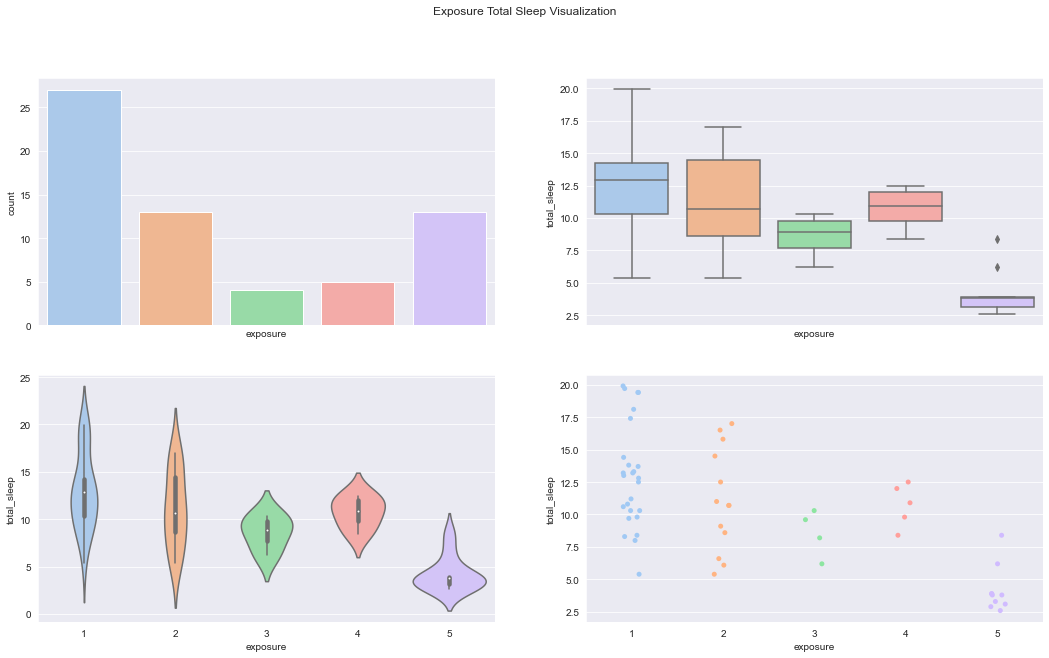
figure, axes = plt.subplots(2, 2, sharex=True, figsize=(18,10))
figure.suptitle('Danger Total Sleep Visualization')
sns.countplot(x='danger',data=df,palette='pastel', ax=axes[0][0])
sns.boxplot(x="danger", y="total_sleep", data=df, palette='pastel', ax=axes[0][1])
sns.violinplot(x="danger", y="total_sleep", data=df, palette='pastel', ax=axes[1][0])
sns.stripplot(x="danger", y="total_sleep", data=df,jitter=True, palette='pastel', ax=axes[1][1])C:\Users\dghr201\AppData\Local\Temp\ipykernel_42120\3554697531.py:6: FutureWarning: Passing `palette` without assigning `hue` is deprecated.
sns.stripplot(x="danger", y="total_sleep", data=df,jitter=True, palette='pastel', ax=axes[1][1])<AxesSubplot:xlabel='danger', ylabel='total_sleep'>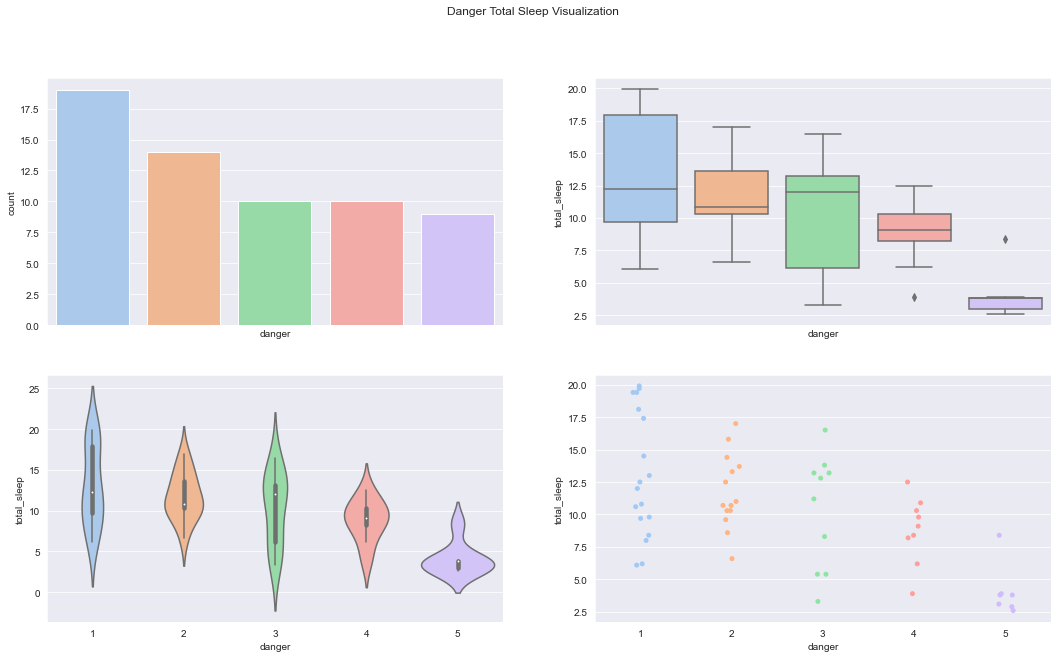
x = explanatory / independent variables y = response / dependent variable